|
|

For Chemistry Professor Steven Benner, Life As We Know It May Not Be The Only AlternativeBy Carolyn Gramling Steven Benner is a science fiction writer’s dream. Science fiction uses present-day facts to speculate about possible futures, envisaging consequences and seeking solutions. Novelist Ray Bradbury once said that the history of mankind is nothing but science fiction: transforming dreams into reality. Over the past 20 years, Benner’s research has examined the limits of scientific possibility. He has pondered how life might exist in seemingly uninhabitable environments, created a flexible new strand of DNA and reconstructed the chemical conditions for the origins of life on Earth. But like the themes of classic science fiction novels, the underlying questions are fundamental and familiar: they center on the chemical basis of life. “We’re really trying to understand what we can do with this pictorial representation of reality on a tiny scale,” Benner says, holding up a colorful, Tinkertoy-type model of DNA. Chemistry, he says, has been taken for granted in recent years. Science has leapt forward from the groundbreaking discovery of the structure of DNA to mapping out the genomes, or DNA blueprints, of a multitude of Earth species from viruses to plants to mammals. “There’s this notion that we can understand the behavior of the world around us in terms of the behavior of molecules and their interactions,” he adds. “But there’s no reason to believe that the answer is entirely written in chemical structure. “What can we do with that theory? Where can we advance it? Where are those advances that open up doors and let you do things technologically that you couldn’t do before?”
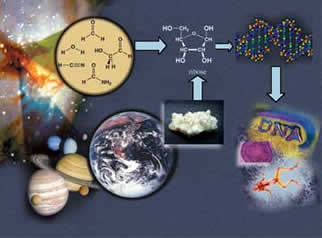
In January 2005, the international Cassini-Huygens mission to Saturn’s moon Titan made landfall. The only moon in the solar system with its own atmosphere, Titan is one place where scientists might find signs of extraterrestrial life — and though it may sound fantastic, that’s not science fiction, Benner says. But the difficulty is in knowing what to look for. “You ask a question about what kinds of molecular systems would support life on Titan — you’re already into the crackpot realm,” Benner says. “But if we’re going to go to Titan, there is very much a need to decide, once we land there, if we’ll recognize the life that might be present.” That’s the job of the National Academy of Sciences’ “weird life” committee, a panel of 12 leading scientists, including Benner, whosemission is to brainstorm where and how such extraterrestrial life might exist. More officially known as the Limits of Organic Life of Planetary Systems, the committee was created in September 2004 and is chaired by John Baross, an astrobiologist at the University of Washington. “Steve is the key player in this whole thing,” Baross says of the committee. “This wouldn’t happen without Steve. He’s a superb organic chemist and he thinks outside the box. He thinks a lot about weird organic chemistry, more than probably anybody on Earth.” The first step in finding weird life is to tilt the focus of the search away from what is considered normal and expected on Earth, Benner says. This is particularly true in the case of water, which was once thought to be an absolute prerequisite to life. That is now considered an Earth-centric point of view. However, organic chemists do believe that having a solvent — a liquid that dissolves other substances, like the broth in the primordial soup — is certainly a helpful part of the process. But there’s no reason that liquid must be water, Benner says. That kind of unconventional thinking is the primary assignment of the weird life committee, particularly as it sets about the task of analyzing the information from the Titan mission. New and clever ideas are essential to its success. “Steve’s excited about this project, and that’s the key thing,” Baross says. “It means he’s actively thinking about it all the time.” Only 50 years ago, scientists just wanted to figure out how DNA was put together. DNA, as James Watson and Francis Crick discovered in 1953, is a ladder-shaped molecule, a double helix composed of two parallel strands connected by many “rungs” made up of four nucleotides, or amino acid bases, arranged in pairs. The order of those four base pairs is unique to each organism and contains its individual code, the genetic instructions for life written as a string of letters — G, A, T and C, representing guanine, adenine, thymine and cytosine. “When the DNA model was introduced it was obviously correct,” Benner says. “Everyone said, ‘Of course, that’s how it has to be.’ But the problem is, if you look at it closely, it doesn’t have to be that way.” With that in mind, Benner has pioneered an emerging field known as “synthetic biology,” a discipline that encompasses genetic engineering and nanotechnology and opens the door to the possibility of Jetson-ish marvels like personalized, genetically tailored medicine. In 2004, the journal Nature featured an article on Benner and synthetic biology, which has been simultaneously heralded for its prospects in biomedicine and environmental engineering and feared for its potential dangers. But though the risks are great, so are the potential rewards. Benner’s research in synthetic biology began while an assistant professor in Harvard’s chemistry department. In 1984, his laboratory pioneered protein engineering, creating a gene that was able to encode an enzyme. That discovery paved the way for future genetic engineering. After leaving Harvard in 1985, Benner spent the next 12 years at the Swiss Federal Institute of Technology, where he began to examine ways to build on Watson and Crick’s work by actually altering DNA’s structure. In 1989, his lab developed a synthetic molecule that mimicked DNA — but had a more flexible framework than the original molecule, enabling it to incorporate more than the four naturally occurring bases. Benner’s molecule could incorporate an additional 12 synthesized bases, or six pairs, in its structure. More turned out to be better, in the case of DNA. With more bases, the possibilities for pairing them also increased, giving biotechnologists enormously enhanced control over how the DNA strands could be assembled. This synthesized molecule is now the basis for a highly sensitive diagnostic tool for doctors, who use it to measure the loads of resistant strains of viruses like hepatitis C and HIV in patients. Because the synthetic bases are inert, don’t interact with natural DNA and occur only in the designed molecule, they act like “tags,” allowing scientists to create highly specific strands of the molecule that can easily be detected and traced. Since joining UF’s Department of Chemistry in 1997, Benner has improved on his own creation. In early 2004, he designed the first artificial DNA-like molecule that was able to copy itself, and then make copies of the copies. That molecular “photocopying” operation, or polymerase chain reaction, is similar to natural Darwinian evolution. To create such a molecule capable of reproducing itself, the researchers had to find an enzyme, known as a DNA polymerase, which would hold the GATC building blocks of natural DNA in the positions necessary to create the famous Watson-Crick base pair. They then assembled the correctly paired nucleotides into a strand.
“DNA polymerases have evolved for billions of years to accept the four natural letters in DNA — A, T, C and G,” Benner says. “Coaxing them to accept two new letters, like K and X, was difficult.” Artificial DNA that can reproduce itself has improved potential to flag disease-related DNA from unwanted viruses, bacteria and possibly biological warfare agents, Benner says, and also helps in the detection of natural genetic defects that cause cancers and genetic diseases. “Our artificial DNA has widespread benefit for patients in diagnostics,” he says. “But until now, it has been largely passive. It has not been able to copy itself.” In October 2004, the National Science Foundation recognized this emerging field by awarding Benner, with colleagues Jack Szostak of Harvard and Gerald Joyce of the Scripps Research Institute, $1.5 million over a three-year period to create a Chemical Bonding Center. The center’s stated objective is to study the origin and evolution of life, and to direct that knowledge to improve the human condition — with the long-term goal of making artificial chemical systems that can self-replicate and thereby evolve. One possible way to improve the human condition, Benner believes, is through a “personalized medicine” system, tailored to each person’s individual genetic code. He is currently developing the tools to make this possible. A prerequisite for personalized medicine is affordable human genome sequencing — currently, the cost for sequencing the 3 billion letters of the human genome is at least $10 million. The target price, per genome, is in the neighborhood of $10,000, he says. But that’s not as far away as it might seem. In 2003, the National Human Genome Research Institute, part of the National Institutes of Health, awarded an $11 million grant to create a Center of Excellence in Genomic Science located jointly at UF and at Columbia University, with UF professors Benner, Leonid Moroz and Weihong Tan as faculty investigators. One stated goal of the center is to develop a range of new tools to do systems biology, and to apply it to the nervous system. And in October 2004, Benner was awarded a $1.6 million, three-year grant by the NIH to develop inexpensive, high-throughput DNA analysis tools to support personalized medicine. The research, done in part with UF chemistry Professor Charles Martin, aims to reduce the cost of sequencing the human genome to $1,000. “These kinds of technology are going to allow you to sequence your genome cheaply,” he says. “A lot of how patients react to a drug is genetically determined. We need to be able to go in and profile you as a patient and decide what medication you’re going to do well with, and what you’re not going to do well with.” “In medicine, the primary escalating costs are labor — which is largely doing complicated diagnostics — followed by treatment,” Benner says. “Obviously, the more predictive medicine we can do, the more we cut the costs. You’ve got to be able to understand a little bit more about individual humans as they interact with the medical system, to see how they’re going to respond.” Other clues for using the sequence of the human genome to diagnose and treat diseases may lie in our distant past, Benner believes. “The past is key to the present,” he says. “By understanding how proteins evolved together with the history of the planet, we can better understand how proteins work today.” Proteins control life functions. To find how they evolved, molecular geneticists extract information about their ancestors by matching up similarities in gene sequences, which provide the recipes for proteins. The process is similar in historical linguistics, which reconstructs ancient languages by finding similarities in their descendent languages. For example, in many European languages, the word for “snow” contains an “n,” which suggests the words stem from another word in a common Indo-European language containing that letter. Instead of languages, geneticists seek similarities in amino acids denoted by the different letters of the genetic code. They use these similarities to reconstruct the amino acid sequences of ancient proteins and resurrect these ancient proteins in the laboratory, studying them to gain insights about their modern descendants. Benner’s work reconstructing the dates of the events that produced the proteins was the subject of a 2002 paper published in the journal Science. With the dates in hand, scientists can match the events that created genes and proteins with events recorded in the fossil and geological records. This allows scientists to zero in on the genes and proteins they may be hunting, he says. Scientists have reasonable knowledge of the biological, chemical and physical nature of life currently on Earth, Benner says. “But we don’t know enough about behaviors of molecules in general to know whether that solution is unique, and no other chemical solution would do, or whether there are other closely related solutions that might happen on Mars or Titan. Or whether there are some ridiculously different solutions like you see on Star Trek.” By closely examining the images of Titan’s rocky surface and ancient lakebeds from the Cassini-Huygens mission, scientists may get closer to some of those answers than ever before. Titan is dominated by nitrogen, methane and other organic, or carbon-based molecules — conditions that are believed to resemble those of the earliest days of the Earth 4.6 billion years ago. It could therefore tell scientists more about the kind of chemical reactions that made it possible for life to emerge on Earth. “It’s a hot topic right now, a topic that we just don’t have a lot of information on,” says Baross. New information from Titan suggests the moon may have had a lake at some point, possibly of methane. The committee met again in March to go over the data from Titan in detail, to think ahead to future explorations and to the search for life on planets with even more apparently inhospitable environments.
“This panel is one of the most interesting things I’ve done,” Benner says. “Because we’ve got people saying, could there be life in concentrated sulfuric acid? Well, why do you ask? Because sulfuric acid is what the atmosphere is above Venus. Okay, could there be life in supercritical hydrogen/helium? Hydrogen is the predominant solvent in the solar system’s gas giants — Jupiter, Saturn, Uranus and Neptune. They’re all made out of it. So could there be life in that? “Until we do the exploration, it’s purely a matter of faith,” he adds. “It depends on whether you believe that life is an intrinsic property of organic molecules.”
Steven Benner |